
Published on the 19th March 2018 by ANSTO Staff
Membrane proteins play a vital role in many critical biological processes. Nearly 30% of proteins in eukaryotic cells are known to be membrane proteins. Mutations or improper folding of these proteins is associated with many known diseases such heart disease, cystic fibrosis, depression, obesity, cancer and many others.
Currently approximately 60% of available drugs target membrane proteins. Despite this, membrane proteins are underrepresented among atomic structures solved, making up less than 1% of the structures in the Protein Data Bank (PDB). This is not due to a lack of biological importance but to the technical challenges associated with their structural analysis and structural determination. Detergents are commonly used to stabilise membrane proteins in order to study their structures.
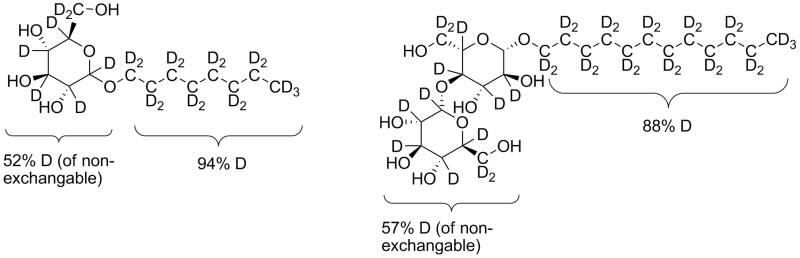
The Chemical Deuteration team of the National Deuteration Facility has developed a new method for the synthesis of common detergents octyl β-D-glucopyranoside (OG) and n-dodecyl-β-D-maltopyranoside (DDM) that allows for selective deuteration such that they had the same neutron scattering length density as 100% deuterium oxide (D2O). This way, only the signal from the membrane protein remained in the small angle neutron scattering (SANS) data.
Researchers from The University of Copenhagen have demonstrated the applicability of this method to five very different membrane proteins using SANS on the QUOKKAinstrument at the Australian Centre for Neutron Scattering and other neutron scattering instruments at overseas facilities. The controlled deuterium isotope labelling of detergents makes solution structure determination of membrane proteins by SANS and subsequent data analysis available to nonspecialists, and it is anticipated that this method will allow rapid screening of membrane proteins structures which have been previously disabled due to the technical difficulties of the experiments.
 |
| Deuterated detergents OG (left) and DDM (right) |
The full publication can be found here: http://www.doi.dx/10.1111/febs.14345
Reference: Midtgaard, S. R.; Darwish, T. A.; Pedersen, M. C.; Huda, P.; Larsen, A. H.; Jensen, G. V.; Kynde, S. A. R.; Skar‐Gislinge, N.; Nielsen, A. J. Z.; Olesen, C.; Blaise, M.; Dorosz, J. J.; Thorsen, T. S.; Venskutonytė, R.; Krintel, C.; Møller, J. V.; Frielinghaus, H.; Gilbert, E. P.; Martel, A.; Kastrup, J. S.; Jensen, P. E.; Nissen, P.; Arleth, L. The FEBS Journal2018, 285, 357.