
Published on the 29th September 2023 by ANSTO Staff
Key Points
-
Research has revealed how the shape of a key protein in the rabies virus contributes to its properties
-
By characterising challenging, intrinsically disordered viral proteins structurally, a large research team provided insight into how the rabies virus can survive hostile environments for long periods of time
-
Small angle X-ray scattering on the SAXS beamline and a number of other techniques, such as mass spectrometry and solution NMR, advanced an understanding of the various roles of isoforms in the activity of the virus
A large team of international researchers have used techniques at ANSTO's Australian Synchrotron to understand how key proteins contribute to the virulence of the rabies virus, sometimes called the 'zombie virus.'
Dr Ashish Sethi, instrument scientist and Acting Scattering Group Manager at the Australian Synchrotron and part of a large international team of scientific experts, said the group wanted to investigate the role of a specific subgroup of viral proteins in virulent and weak strains of rabies by studying it at the molecular level.
Rabies virus (RABV) has only five genes, but it manages to do a lot with its limited genetic material. It achieves this by producing five different versions of a particular protein called the phosphoprotein (P protein) from its P gene. These different versions of the P protein, known as isoforms, have various roles in the activity of the virus.
P3 proteins have differences in their ability to combat their hosts’ immune system, move into the cell nucleus, and associate with structures called microtubules, which are important in the ability of the virus to cause disease.
These challenging, intrinsically disordered viral proteins were structurally characterised using an integrated structural biology approach which includes small angle X-ray scattering on the SAXS beamline at the Australian Synchrotron and a number of other techniques, such as mass spectrometry and solution NMR.
“The characterisation provided insight into how the rabies virus can survive hostile environments for long periods of time,” said Dr Sethi.
These techniques also provided insights into the structure of the protein, which could help with the development of better therapeutics for rabies,” he added.
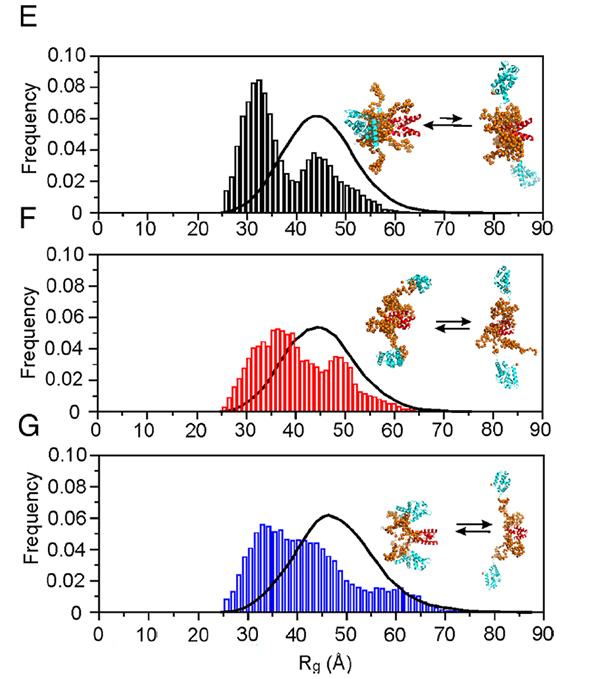
The research, published in PNAS and featured in an edition of Australian Biochemist, revealed that the shape of the P3 protein, open or closed, contributed to its diverse properties.
The P3 protein in the disease-causing strain can change between the two shapes, while the P3 protein in the weakened/avirulent strain stays mostly in the open shape, highlighting how critical these conformations are.
These interactions between domains and distant disordered parts of the P3 protein likely play a role in its various functions.
“Like the Australian bat lyssavirus, rabies has a small genome expressing five proteins – including the crucial Phosphoprotein (P protein), which is conserved for replication and survival.
“The coding capacity of the P protein is increased as it expresses five isoforms, which, when truncated, results in an extensive gain of new functions,” explained Dr Sethi.
“The greater the shortening, the greater the functional diversity, which is so unique and fascinating,” Dr Sethi explained.
“We have shown that going from one isoform, P1, down to the third isoform (P3) results in the gain of a significant subset of new functions, and that explains why the virus needs these different isoforms.”
“The truncated P3 isoform demonstrated a gain in function, including an enhanced capacity to migrate into the nucleus to play with their host's genetic response, which results in immune evasion.
“This research advances an understanding of multifunctionality and indicates that higher-order organisation and structural flexibility is critical to the unique gain-of-function by the P3 isoform.”
Other contributing institutions included Monash University, Dana-Farber Cancer Institute Harvard University, University of Melbourne, and Gifu University (Japan).
According to Dr Sethi, the research will be supported in the future by the operation of the new high-flux BioSAXS Beamline.
“Because there will be so many photons … we won’t really need a large quantity of protein samples. The new beamline will allow us to visualise additional and unique conformations of disordered protein isoforms, and I can’t wait to record the first protein SAXS experiment on our new BioSAXS beamline,” he said.